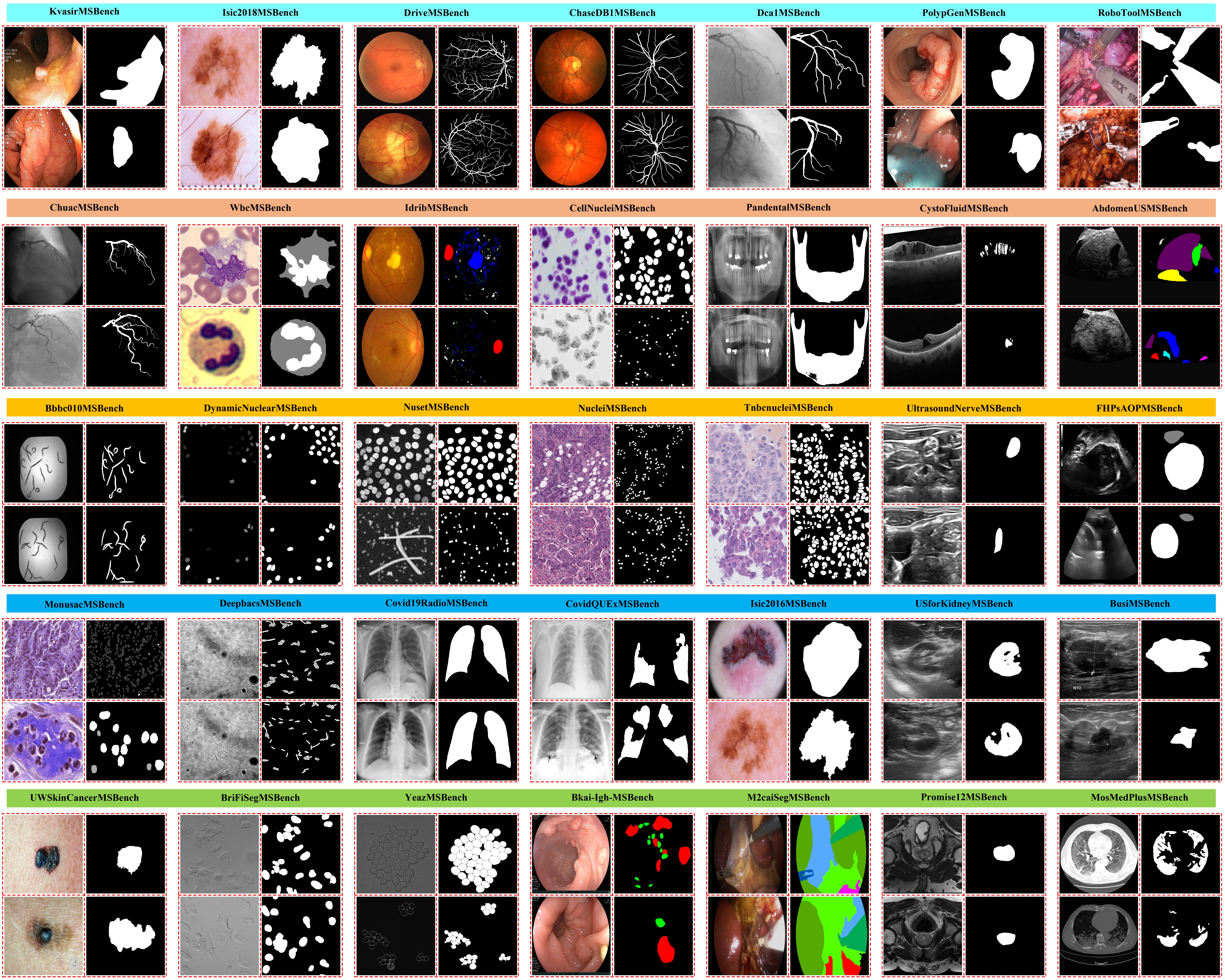
MedSegBench is a comprehensive benchmark designed to evaluate deep learning models for medical image segmentation across a wide range of modalities. It covers a wide range of modalities, including 35 datasets with over 60,000 images from ultrasound, MRI, and X-ray. The benchmark addresses challenges in medical imaging by providing standardized datasets with train/validation/test splits, considering variability in image quality and dataset imbalances. The benchmark supports binary and multi-class segmentation tasks with up to 19 classes and uses the U-Net architecture with various encoder/decoder networks such as ResNets, EfficientNet, and DenseNet for evaluations. MedSegBench is a valuable resource for developing robust and flexible segmentation algorithms and allows for fair comparisons across different models, promoting the development of universal models for medical tasks. It is the most comprehensive study among medical segmentation datasets. The datasets and source code are publicly available, encouraging further research and development in medical image analysis.
Setup the required environments and install `medsegbench` as a standard Python package from PyPI:
pip install medsegbench
Or install from source:
pip install --upgrade git+https://github.com/zekikus/MedSegBench.git
You can use default 512 sized version using the downloaded files:
>>> from medsegbench import Promise12MSBench
>>> train_dataset = Promise12MSBench(split="train")
You can download the dataset by setting `download=True`:
>>> from medsegbench import Promise12MSBench
>>> train_dataset = Promise12MSBench(split="train", download=True)
You can download different sized versions by setting `size={128, 256, 512}`:
>>> from medsegbench import Promise12MSBench
>>> train_dataset = Promise12MSBench(split="train", size=256)
You can download sub-categories of dataset by setting `category={C1, C2, C3, ...}`:
>>> from medsegbench import WbcMSBench
>>> train_dataset = WbcMSBench(split="train", category='C1')
| Dataset Name | Modality | Pathology/Organ Studied | Binary or Multi-class (# Classes) | # Images | # Train/Val/Test |
|---|---|---|---|---|---|
| AbdomenUSMSBench | Ultrasound | Gallbladder, Kidney, Liver, Spleen, Vessel | Multi-class (9) | 926 | 569/64/293 |
| Bbbc010MSBench | Microscopy | Caenorhabditis elegans | Binary | 100 | 70/10/20 |
| Bkai-Igh-MSBench | Endoscopy | Colon polyps | Multi-class (3) | 1,000 | 700/100/200 |
| BriFiSegMSBench | Microscopy | Lung, Cervix, Breast, Eye | Binary | 1,360 | 1005/115/240 |
| BusiMSBench | Ultrasound | Breast | Binary | 647 | 452/64/131 |
| CellnucleiMSBench | Nuclei | Nuclei | Binary | 670 | 469/67/134 |
| ChaseDB1MSBench | Fundus | Eye (Retinal vessels) | Binary | 28 | 19/2/7 |
| ChuacMSBench | Fundus | Eye (Retinal vessels) | Binary | 30 | 21/3/6 |
| Covid19RadioMSBench | Chest X-Ray | Lung | Binary | 21,165 | 14,814/2,115/4,236 |
| CovidQUExMSBench | Chest X-Ray | Lung | Binary | 2,913 | 1,864/466/583 |
| CystoFluidMSBench | OCT | Eye (Cystoid macular edema) | Binary | 1,006 | 703/101/202 |
| Dca1MSBench | Fundus | Eye (Retinal vessels) | Binary | 134 | 93/13/28 |
| DeepbacsMSBench | Microscopy | Bacterial cells | Binary | 34 | 17/2/15 |
| DriveMSBench | Fundus | Eye (Retinal vessels) | Binary | 40 | 18/2/20 |
| DynamicNuclearMSBench | Nuclear Cell | Nuclear Cells | Binary | 7,084 | 4,950/1,417/717 |
| FHPsAOPMSBench | Ultrasound | Fetal head, pubic symphysis | Multi-class (3) | 4,000 | 2,800/400/800 |
| IdribMSBench | Fundus | Eye (Optic discs) | Binary | 80 | 47/6/27 |
| Isic2016MSBench | Dermoscopy | Skin (Lesions) | Binary | 1,279 | 810/90/379 |
| Isic2018MSBench | Dermoscopy | Skin (Lesions) | Binary | 3,694 | 2,594/100/1,000 |
| KvasirMSBench | Endoscopy | Gastrointestinal polyps | Binary | 1,000 | 700/100/200 |
| M2caiSegMSBench | Endoscopy | Surgical tools and abdominal tissues | Multi-class (19) | 614 | 245/307/62 |
| MonusacMSBench | Pathology | Lung, Prostate, Kidney, and Breast | Multi-class (6) | 310 | 188/21/101 |
| MosMedPlusMSBench | CT | Lung | Binary | 2,729 | 1,910/272/547 |
| NucleiMSBench | Pathology | Cell Nuclei | Binary | 141 | 98/14/29 |
| NusetMSBench | Nuclear Cell | Nuclear cells | Binary | 3,408 | 2,385/340/683 |
| PandentalMSBench | X-Ray | Mandible | Binary | 116 | 81/11/24 |
| PolypGenMSBench | Endoscopy | Colon polyps | Binary | 1,412 | 984/140/288 |
| Promise12MSBench | MRI | Prostate | Binary | 1,473 | 1,031/147/295 |
| RoboToolMSBench | Endoscopy | Surgical tools | Binary | 500 | 350/50/100 |
| TnbcnucleiMSBench | Pathology | Nuclei in histopathology images | Binary | 50 | 35/5/10 |
| UltrasoundNerveMSBench | Ultrasound | Neck (Brachial Plexus Nerves) | Binary | 2,323 | 1,651/223/449 |
| USforKidneyMSBench | Ultrasound | Kidney | Binary | 4,586 | 3,210/458/918 |
| UWSkinCancerMSBench | Dermoscopy | Skin (Cancer) | Binary | 206 | 143/19/44 |
| WbcMSBench | Microscopy | White Blood Cell | Multi-class (3) | 400 | 280/40/80 |
| YeazMSBench | Microscopy | Yeast Cells | Binary | 707 | 360/96/251 |
| Precision (PREC) | Recall (REC) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | RN-18 | RN-50 | EN | MN-v2 | DN-121 | MVT | RN-18 | RN-50 | EN | MN-v2 | DN-121 | MVT |
| AbdomenUSMSB | 0.976 | 0.973 | 0.950 | 0.964 | 0.955 | - | 0.652 | 0.654 | 0.670 | 0.655 | 0.671 | - |
| Bbbc010MSB | 0.919 | 0.926 | 0.918 | 0.918 | 0.922 | - | 0.912 | 0.909 | 0.904 | 0.900 | 0.920 | - |
| Bkai-Igh-MSB | 0.983 | 0.961 | 0.939 | 0.944 | 0.952 | 0.983 | 0.563 | 0.625 | 0.705 | 0.737 | 0.642 | 0.563 |
| BriFiSegMSB | 0.812 | 0.816 | 0.812 | 0.803 | 0.817 | - | 0.873 | 0.886 | 0.882 | 0.861 | 0.898 | - |
| BusiMSB | 0.729 | 0.753 | 0.765 | 0.766 | 0.794 | - | 0.727 | 0.665 | 0.728 | 0.672 | 0.714 | - |
| CellnucleiMSB | 0.924 | 0.920 | 0.913 | 0.901 | 0.927 | 0.928 | 0.882 | 0.886 | 0.894 | 0.872 | 0.898 | 0.883 |
| ChaseDB1MSB | 0.788 | 0.789 | 0.780 | 0.794 | 0.793 | 0.774 | 0.733 | 0.738 | 0.725 | 0.703 | 0.739 | 0.705 |
| ChuacMSB | 0.713 | 0.710 | 0.643 | 0.644 | 0.870 | - | 0.470 | 0.451 | 0.526 | 0.458 | 0.444 | - |
| Covid19RadioMSB | 0.991 | 0.991 | 0.991 | 0.991 | 0.992 | - | 0.990 | 0.990 | 0.991 | 0.991 | 0.991 | - |
| CovidQUExMSB | 0.741 | 0.738 | 0.753 | 0.739 | 0.760 | - | 0.824 | 0.810 | 0.815 | 0.827 | 0.826 | - |
| CystoFluidMSB | 0.889 | 0.870 | 0.874 | 0.879 | 0.888 | 0.874 | 0.848 | 0.872 | 0.856 | 0.844 | 0.851 | 0.865 |
| Dca1MSB | 0.776 | 0.788 | 0.775 | 0.781 | 0.801 | - | 0.757 | 0.757 | 0.740 | 0.732 | 0.740 | - |
| DeepbacsMSB | 0.957 | 0.956 | 0.955 | 0.958 | 0.959 | - | 0.905 | 0.907 | 0.897 | 0.886 | 0.900 | - |
| DriveMSB | 0.817 | 0.789 | 0.799 | 0.811 | 0.827 | 0.784 | 0.756 | 0.790 | 0.748 | 0.750 | 0.751 | 0.784 |
| DynamicNuclearMSB | 0.924 | 0.929 | 0.937 | 0.926 | 0.928 | - | 0.965 | 0.965 | 0.966 | 0.963 | 0.965 | - |
| FHPsAOPMSB | 0.962 | 0.964 | 0.964 | 0.965 | 0.961 | - | 0.960 | 0.951 | 0.956 | 0.955 | 0.959 | - |
| IdribMSB | 0.150 | 0.153 | 0.139 | 0.150 | 0.172 | 0.110 | 0.089 | 0.072 | 0.065 | 0.078 | 0.068 | 0.041 |
| Isic2016MSB | 0.890 | 0.897 | 0.912 | 0.912 | 0.913 | 0.897 | 0.907 | 0.910 | 0.919 | 0.901 | 0.905 | 0.917 |
| Isic2018MSB | 0.838 | 0.839 | 0.857 | 0.864 | 0.878 | 0.854 | 0.911 | 0.907 | 0.923 | 0.908 | 0.896 | 0.907 |
| KvasirMSB | 0.816 | 0.770 | 0.839 | 0.842 | 0.874 | 0.644 | 0.768 | 0.755 | 0.860 | 0.780 | 0.804 | 0.697 |
| M2caiSegMSB | 0.737 | 0.756 | 0.801 | 0.762 | 0.759 | 0.794 | 0.224 | 0.225 | 0.228 | 0.225 | 0.230 | 0.227 |
| MonusacMSB | 0.945 | 0.951 | 0.951 | 0.951 | 0.951 | 0.951 | 0.589 | 0.589 | 0.589 | 0.589 | 0.589 | 0.589 |
| MosMedPlusMSB | 0.816 | 0.817 | 0.807 | 0.821 | 0.826 | 0.808 | 0.786 | 0.802 | 0.796 | 0.793 | 0.798 | 0.767 |
| NucleiMSB | 0.250 | 0.233 | 0.223 | 0.199 | 0.225 | 0.196 | 0.394 | 0.395 | 0.449 | 0.281 | 0.479 | 0.481 |
| NusetMSB | 0.949 | 0.950 | 0.953 | 0.950 | 0.953 | - | 0.951 | 0.951 | 0.951 | 0.952 | 0.952 | - |
| PandentalMSB | 0.956 | 0.955 | 0.952 | 0.945 | 0.965 | - | 0.967 | 0.968 | 0.963 | 0.958 | 0.965 | - |
| PolypGenMSB | 0.763 | 0.739 | 0.783 | 0.824 | 0.794 | 0.557 | 0.584 | 0.538 | 0.684 | 0.582 | 0.632 | 0.570 |
| Promise12MSB | 0.911 | 0.900 | 0.900 | 0.903 | 0.909 | - | 0.903 | 0.896 | 0.902 | 0.905 | 0.906 | - |
| RoboToolMSB | 0.878 | 0.874 | 0.893 | 0.885 | 0.905 | 0.885 | 0.854 | 0.864 | 0.867 | 0.835 | 0.868 | 0.893 |
| TnbcnucleiMSB | 0.813 | 0.834 | 0.748 | 0.772 | 0.819 | 0.746 | 0.758 | 0.760 | 0.762 | 0.770 | 0.770 | 0.797 |
| UltrasoundNerveMSB | 0.799 | 0.801 | 0.779 | 0.786 | 0.798 | - | 0.796 | 0.782 | 0.814 | 0.791 | 0.802 | - |
| USforKidneyMSB | 0.979 | 0.979 | 0.981 | 0.980 | 0.980 | - | 0.980 | 0.978 | 0.982 | 0.980 | 0.980 | - |
| UWSkinCancerMSB | 0.920 | 0.925 | 0.928 | 0.939 | 0.926 | 0.930 | 0.857 | 0.829 | 0.882 | 0.857 | 0.839 | 0.872 |
| WbcMSB | 0.961 | 0.962 | 0.965 | 0.959 | 0.963 | 0.966 | 0.966 | 0.966 | 0.968 | 0.963 | 0.970 | 0.969 |
| YeazMSB | 0.935 | 0.931 | 0.936 | 0.931 | 0.934 | - | 0.974 | 0.979 | 0.971 | 0.977 | 0.978 | - |
| F1-Score (F1) | Intersection over Union (IOU) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | RN-18 | RN-50 | EN | MN-v2 | DN-121 | MVT | RN-18 | RN-50 | EN | MN-v2 | DN-121 | MVT |
| AbdomenUSMSB | 0.642 | 0.640 | 0.640 | 0.635 | 0.643 | - | 0.632 | 0.630 | 0.628 | 0.624 | 0.632 | - |
| Bbbc010MSB | 0.915 | 0.917 | 0.910 | 0.908 | 0.920 | - | 0.844 | 0.848 | 0.837 | 0.833 | 0.854 | - |
| Bkai-Igh-MSB | 0.554 | 0.617 | 0.692 | 0.733 | 0.630 | 0.554 | 0.546 | 0.604 | 0.676 | 0.713 | 0.615 | 0.546 |
| BriFiSegMSB | 0.826 | 0.834 | 0.831 | 0.816 | 0.840 | - | 0.717 | 0.728 | 0.724 | 0.702 | 0.738 | - |
| BusiMSB | 0.674 | 0.632 | 0.711 | 0.655 | 0.695 | - | 0.578 | 0.547 | 0.624 | 0.565 | 0.615 | - |
| CellnucleiMSB | 0.889 | 0.892 | 0.894 | 0.880 | 0.907 | 0.891 | 0.822 | 0.827 | 0.830 | 0.815 | 0.838 | 0.826 |
| ChaseDB1MSB | 0.758 | 0.761 | 0.750 | 0.744 | 0.764 | 0.735 | 0.611 | 0.615 | 0.601 | 0.594 | 0.618 | 0.582 |
| ChuacMSB | 0.487 | 0.451 | 0.499 | 0.462 | 0.522 | - | 0.357 | 0.334 | 0.369 | 0.340 | 0.400 | - |
| Covid19RadioMSB | 0.991 | 0.990 | 0.991 | 0.991 | 0.992 | - | 0.982 | 0.981 | 0.983 | 0.982 | 0.983 | - |
| CovidQUExMSB | 0.740 | 0.734 | 0.744 | 0.742 | 0.756 | - | 0.627 | 0.620 | 0.633 | 0.631 | 0.647 | - |
| CystoFluidMSB | 0.852 | 0.857 | 0.849 | 0.842 | 0.853 | 0.855 | 0.759 | 0.765 | 0.754 | 0.747 | 0.761 | 0.763 |
| Dca1MSB | 0.762 | 0.767 | 0.753 | 0.751 | 0.765 | - | 0.618 | 0.625 | 0.606 | 0.604 | 0.623 | - |
| DeepbacsMSB | 0.930 | 0.931 | 0.925 | 0.921 | 0.929 | - | 0.869 | 0.870 | 0.860 | 0.853 | 0.867 | - |
| DriveMSB | 0.782 | 0.786 | 0.770 | 0.775 | 0.782 | 0.781 | 0.643 | 0.648 | 0.626 | 0.634 | 0.643 | 0.641 |
| DynamicNuclearMSB | 0.941 | 0.942 | 0.948 | 0.940 | 0.942 | - | 0.895 | 0.897 | 0.906 | 0.893 | 0.897 | - |
| FHPsAOPMSB | 0.961 | 0.957 | 0.959 | 0.959 | 0.960 | - | 0.929 | 0.923 | 0.927 | 0.927 | 0.928 | - |
| IdribMSB | 0.100 | 0.090 | 0.078 | 0.092 | 0.089 | 0.053 | 0.061 | 0.054 | 0.046 | 0.056 | 0.054 | 0.030 |
| Isic2016MSB | 0.878 | 0.887 | 0.903 | 0.891 | 0.893 | 0.891 | 0.803 | 0.814 | 0.836 | 0.820 | 0.825 | 0.822 |
| Isic2018MSB | 0.849 | 0.849 | 0.868 | 0.865 | 0.861 | 0.853 | 0.761 | 0.762 | 0.790 | 0.783 | 0.785 | 0.773 |
| KvasirMSB | 0.739 | 0.698 | 0.812 | 0.754 | 0.794 | 0.569 | 0.645 | 0.596 | 0.733 | 0.668 | 0.718 | 0.457 |
| M2caiSegMSB | 0.214 | 0.215 | 0.218 | 0.216 | 0.223 | 0.217 | 0.190 | 0.191 | 0.196 | 0.192 | 0.200 | 0.194 |
| MonusacMSB | 0.557 | 0.559 | 0.559 | 0.559 | 0.559 | 0.538 | 0.540 | 0.540 | 0.540 | 0.540 | 0.540 | 0.540 |
| MosMedPlusMSB | 0.780 | 0.790 | 0.781 | 0.785 | 0.791 | 0.761 | 0.674 | 0.682 | 0.674 | 0.679 | 0.686 | 0.650 |
| NucleiMSB | 0.282 | 0.274 | 0.278 | 0.205 | 0.275 | 0.253 | 0.169 | 0.164 | 0.167 | 0.119 | 0.166 | 0.150 |
| NusetMSB | 0.949 | 0.949 | 0.951 | 0.950 | 0.951 | - | 0.906 | 0.906 | 0.909 | 0.907 | 0.910 | - |
| PandentalMSB | 0.961 | 0.961 | 0.957 | 0.950 | 0.965 | - | 0.926 | 0.926 | 0.919 | 0.907 | 0.932 | - |
| PolypGenMSB | 0.573 | 0.541 | 0.666 | 0.588 | 0.621 | 0.477 | 0.495 | 0.457 | 0.587 | 0.512 | 0.545 | 0.382 |
| Promise12MSB | 0.895 | 0.888 | 0.892 | 0.896 | 0.900 | - | 0.828 | 0.817 | 0.821 | 0.827 | 0.832 | - |
| RoboToolMSB | 0.856 | 0.859 | 0.874 | 0.847 | 0.879 | 0.882 | 0.765 | 0.769 | 0.788 | 0.753 | 0.798 | 0.798 |
| TnbcnucleiMSB | 0.779 | 0.785 | 0.738 | 0.762 | 0.788 | 0.759 | 0.641 | 0.652 | 0.596 | 0.621 | 0.654 | 0.618 |
| UltrasoundNerveMSB | 0.782 | 0.776 | 0.787 | 0.772 | 0.786 | - | 0.671 | 0.664 | 0.675 | 0.660 | 0.676 | - |
| USforKidneyMSB | 0.979 | 0.978 | 0.981 | 0.980 | 0.980 | - | 0.960 | 0.958 | 0.963 | 0.961 | 0.960 | - |
| UWSkinCancerMSB | 0.864 | 0.846 | 0.890 | 0.879 | 0.856 | 0.881 | 0.795 | 0.766 | 0.818 | 0.803 | 0.779 | 0.813 |
| WbcMSB | 0.962 | 0.963 | 0.966 | 0.959 | 0.966 | 0.967 | 0.930 | 0.931 | 0.937 | 0.926 | 0.936 | 0.938 |
| YeazMSB | 0.953 | 0.953 | 0.952 | 0.952 | 0.954 | - | 0.912 | 0.912 | 0.909 | 0.910 | 0.914 | - |
The MedSegBench dataset is licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0), except CystoFluidMSBench, CovidQUExMSBench, MonusacMSBench under Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0), USforKidneyMSBench under GPL2, DynamicNuclearMSBench under (modified Apache license). Furthermore, all datasets can be used for both academic and non-commercial purposes. Users who utilize the datasets within Medsegbench must also comply with the license terms specified for these datasets (see license in the info.py).
The code under Apache-2.0 License
@article{Ku2024,
title = {MedSegBench: A comprehensive benchmark for medical image segmentation in diverse data modalities},
volume = {11},
ISSN = {2052-4463},
url = {http://dx.doi.org/10.1038/s41597-024-04159-2},
DOI = {10.1038/s41597-024-04159-2},
number = {1},
journal = {Scientific Data},
publisher = {Springer Science and Business Media LLC},
author = {Kuş, Zeki and Aydin, Musa},
year = {2024},
month = nov
}Please also cite the corresponding paper(s) of source data if you use any subset of MedSegBench (check this bibtex).